


Intro -prelude from Dan Winter: Academic Paper - CIRCULAR DNA - from Ken Biegeleisen.
Update: Ken Biegeleisen on DNA is not a Helix- references in 2010: www.notahelix.com/
also see Dec 04 - Addition on bottom of this article
-
It is a pleasure to share this RINGING DNA material - thanks to Ken Biegeleisen - who has formalized and represented his paper here on TOROIDAL or CIRCULAR DNA (relating to the concept of 'Braiding' DNA) - with academic references.
Misconception ONE: I am going to get spiritual and cosmic and immortal (biologically sustainable) when divine force (the astrology of charge?) adds '12 strands' to my DNA.
More the Truth: IF you choose the hygiene to eat, and live in, and dance in, and HAVE BLISS in-- - LIFE FORCE and therefore CHARGE - THEN your DNA may absorb that charge by enveloping itself in the recursive braid which ENSOULS - (implodes). The addition of strands which appear to happen when you braid thread into string into rope, is because of braiding (the answer lies 'folded in an envelope' - see pics below). Braiding your DNA is something you do and learn by choice - to absorb 'spin' (charge) - not something 'GOD' does for you. Only the strength of will growing in the sun-shine of shareable and embedable intent - is sustainable (and therefore able to be 'saved').
IF you tilt a cube 32degrees you can spin it into the next dimension (axis of symmetry) which is a DODECA (4th Dimension)..THEN you step or wratchet that dodecahedron down a HELIX or SLINKY (the SHAPE of DNA)- that is the 5th dimension (axis of symmetry). SO the THREAD that begins DNA is 5 spin symmetries or dimension.
FINALLY.. if you take THAT thread and recursively braid the braid of the braid.. (thread into string into rope into fat rope into fatter rope- THIS is how DNA accepts the charge into a re-CORDing of its environment)..
and on the SEVENTH time you recursively braid or plait - PRESTO - the DNA becomes a DONUT / TORUS!. (YOU are RINGlord). This is WHY the seventh spin or the 7 color on the topo map makes DONUT. AT this point you do indeed have 12 AXIS OF SYMMETRY of DIMENSIONS (as opposed to 12 STRANDS ) in your DNA.. AND - the implosion of that donut DNA sending (by recursive adding a phase velocities thru lightspeed ) ALL of the charge symmetry of ALL your biological memory THRU THE SPEED OF LIGHT. And THAT - is what makes a SOUL (the tornado thru light speed )- indicated appropriately by the ability to
+lucid dream
+take memory thru death
+time travel without embarassing heavy metal craft
+make gravity in genes
+steer stars.. etc.
(called Chloridians in STAR WARS - Anikin- -.. also called BOSON SEVEN in the DNA power spectra taken at Montauk to predict WHO could steer the time (Deca Delta / Dodeca - see "Contact" movie) antenna "chair".
The remnant 'fallen Nephalim' conservative DNA culture from Alpha Draconis thru Enlil / Yalweh / Michael / Michabo / Amon - then stablished the rules to PREVENT DNA from getting FREE - which became India's CASTE SYSTEM - and Aboriginal Marriage laws.. and Jewish families who could only make love thru a hole in the sheet. The machine based 'borg' Nephalim culture (lo-oxygen types) doesn't want you to ignite your DNA with BLISS to psychokinesis - because that (lucid dreaming / shamanic) skill is the ONLY technology (the real TRON - see BST ) they have no response to. It takes WILL to implode and ensoul DNA - which no passive GOD IS OUTSIDE YOU parasite priests can create. The force of WILL involves HYGIENE for BLISS.
reference pictorial aritcles:
Letter to the UN on Genetic Engineering as Coherence LOSS now Measureable.
Misconception TWO: The compression spin map called "Alphabet" on the side of the "LORD OF THE RING" - RING - is too powerful for anyone to hold.
More the Truth: The 'RINGing' which causes thoughts / emotions to become objects is the implosive nature of DNA which when braided recursively by BLISS compresses charge in the only process (charge compression) which turns light (ether) into matter. (and DNA into its real destiny as a gravity making star bender) .The index to those possible angles of (Golden Ratio) compression is precisely the equation for the origin of most alphabets of Earth - specifically including Hebrew and Arabic... for the specific reason that only those biological structures which USE that symmetry discipline get to be sustainable (immortal).
Misconception THREE: DNA controls our destiny.
More the Truth: DNA is a shapeshifting mag worm which merely bends itself as a wave around the fields we form by bending the magnetism we call emotion and sacred space. It is a relatively passive mechanical linkage device designed to helpfully slow down the rate at which emotions become objects- so that we FEEL and understand the connection between CREATION and what we just now felt. Once the need for slowing down that response of stars to feeling, no longer exists - then DNA is replaced by its own etheric essence (superluminal compression field) - and we can bend light by implosion directly (only love bends the light / embedding).
If we choose consistent spin-dense and sustainable magnetic environments - then DNA responds by adding that inertia to our life force - so that eventually we gather the inertia to properly create stars and become suns ourselves. Otherwise we die a death which is permanent. DNA is merely a biofeedback device to tell us when we have consciously chosen the CHARGE and LIFE FORCE - which makes us sustainable. Short-circuiting the function of DNA by changing the spin links we call codons mechanically is not only un-sustainable and destructive - but it fundamentally ignores the design of nature - which was that DNA help us take responsibility for the SHAPE OF MAGNETISM - and so learn that bending light into shape is how we too may create stars.
Wouldn't you think that KNOT THEORY could explain WHY the 7th recursive braid of DNA (from BLISS?) renders it Toroidal? Implosive? and Ensouled? and Lucid Dream / Time Travel ready???
per discussion at: goldenmean.info/knotslipping
--
below on Knot Theory from: KnotPlotSite - http://www.pims.math.ca/knotplot/links/sphere.html


 Toroidal Coiling
of DNA - Comparing Observed Data with Computer Models- from: http://www-vis.lbl.gov/Vignettes/KDowning-DNA/
Toroidal Coiling
of DNA - Comparing Observed Data with Computer Models- from: http://www-vis.lbl.gov/Vignettes/KDowning-DNA/
The figure shows three electron microscope images of DNA toroids accompanied by computer simulations of toroids in corresponding orientations. On the top left is a toroid seen nearly perpendicular to the direction of view, so that the ~23 Å spacing between rows of DNA strands, reflecting the hexagonal packing of strands throughout most of the toroid, is clearly seen around almost the entire toroid. The region where the fringe spacing is least visible may correspond to a region where the DNA strands cross over between loops at different radii. In the middle a toroid is seen edge on, revealing the hexagonal packing on the lower side and disordered packing on the top. The toroid in the right-hand image is tilted so that the fringes spacing is seen over only a small part of the toroid. Below each micrograph is a view of a solid model oriented to match the toroid. On the left, the region of crossovers is localized near the 4-o'clock position. The front half of the model in the center is cut away to reveal the hexagonal packing in the lower half and disorder in the crossover region toward the top.
The motivation for this work arises from the academic interest in the behavior of DNA, the fact that DNA is sometimes naturally packed in these toroidal arrays, for example in some sperm and bacteriophages, and the possibility that this may be a useful way to package DNA for genetic therapy.
From: http://www.pnas.org/cgi/content/full/100/16/9296
Within living cells, DNA is highly condensed as compared with
free DNA in solution (1). In sperm cells and viruses, where
gene transcription is inactive, DNA condensation approaches the
limits of molecular compaction (2, 3). The condensation of
free DNA in vitro has long been of interest as a potential model
for DNA condensation in vivo, particularly as a model of DNA packaging
in viruses (46). More recently, DNA condensation has attracted
much attention for its direct relevance to the preparation of
DNA for delivery as a part of gene therapies (710).
More than 25 years ago it was discovered that multivalent cations
cause DNA to collapse from solution into toroid-shaped condensates
Readers of the Dan Winter web site are accustomed to various speculations about the physical, mathematical and spiritual significance of the world-famous Watson-Crick "double-helix" structure of DNA, and it is therefore with some trepidation that I "rain on the parade", by showing why the double-helical structure-pretty picture that it is-is nevertheless probably not the structure of DNA in any living organism.
The primary "business" of DNA is replication. DNA carries the "codes" for the structural proteins and enzymes which give an organism its characteristics, and these codes must be passed from generation to generation.
What exactly gets passed? The codes consist of sequences of nitrogenous bases: adenine, thymine, cytosine and guanine, usually just abbreviated A, T, C, and G. These 4 "letters" are the "alphabet" of genetics, much akin to the binary code which lies at the root of all computer language. Just as a computer ultimately "spells" every program with just two numbers ("0" and "1"), likewise, the code for every structural protein and gene in the human body is "spelled" with an alphabet of only 4 bases in various combinations: A, T, C, and G.
A "gene" is therefore a linear string of bases, like a string of pearls. The bases are held together by a backbone made of sugar and phosphate. In order for the cell to execute the so-called "genetic code", it employs an extraordinary system which reads the strings of bases, and converts the information into corresponding strings of amino acids, which are the building blocks of protein. The fact that a string of bases in DNA codes for a corresponding string of amino acids in protein is sometimes called the "Central Dogma of Molecular Biology" (G-d help us! -- Is this really what we need in the post-Copernican world?).
So much for the genetics. The manner in which the bases are organized on the sugar-phosphate backbone was deduced by Watson & Crick in 1953, and, regardless of what modifications are made to their structure, this deduction will remain as one of the towering milestones in the history of science, as long as science is practiced in the world.
The manner in which the entire DNA molecule is organized, however, is another question altogether. It makes no difference to genetics whether DNA is a right-handed helix, a left-handed helix, or not-a-helix at all. Genetics simply requires that there be a string of bases, the "alphabet" if you will. Whether the sugar-phosphate backbone to which the bases are attached is twisted or not is totally irrelevant.
If this is the case, why then do scientists think that DNA is twisted?
The original structural studies of DNA were done by Maurice Wilkins, who shared the Nobel Prize with Watson & Crick (Rosalind Franklin, who actually did most of the work attributed to Wilkins, was neglected, and is now a major historical figure in the fight for women's equality). Wilkins and Franklin did what's called "x-ray crystallography". This means they took pure DNA and turned it into crystals, then took x-ray pictures of the crystals.
To make crystals, totally pure DNA is required. DNA solutions are gooey, and when a glass rod is dipped into DNA and raised up in the air, the sticky liquid is drawn into a thin thread. If the thread is drawn thin enough, it will dry out and crystallize, and a picture of the crystal can be taken with x-rays. The picture looks sort of like a snowflake. Within this very intriguing geometric pattern lies hidden information about the structure of the crystal. This information must be pulled out by mathematics. The formulae involved are difficult in the extreme, and in the old days, people who actually did the math (called "Fourier Transformation") were held somewhat in awe by other scientists. Nowadays, however, all the mathematical "leg work" is done by computers, and the process has been largely de-mystified.
The picture of DNA which emerged from the Wilkins x-ray studies was the double-helix, and very few people, including disagreeable people like myself, have ever had any reason to doubt that the structure of DNA in artificial crystals is a right-handed double-helix. But is that the structure of DNA in cells? Who can say?
Humans, insofar as our cell type is concerned, are eukaryotes, meaning that our cells are of the type referred to as "higher cells". The "lower cells" are called prokaryotes, and include, for example, bacteria.
Now, humans are not the only eukaryotes. All "higher life forms", both animal and vegetable, are eukaryotes. This means all plants, even including some unicellular plants such as mold and yeast, and all animals, even including some unicellular life forms such as paramecium or amoeba.
It is a known fact that the DNA of all eukaryotic cells is invariably bound to proteins, called histones. Here's an interesting fact for you to contemplate: The histones are exactly the same in all "higher" life forms. That means that the histones of human cells are the same as the histones of the pea plant and the amoeba. Doesn't this mean that histones are critically involved in chromosome structure? Of course it does. There's no such thing as "pure" DNA in cells! In the real world, in life, all DNA is bound to protein. There are no exceptions to this rule.
The structure of pure DNA is said to be known down to the angstrom level, and if you don't know what an angstrom is, let me tell you that it's very, very small. In other words, the structure of DNA is known down to the minutest detail.
Also, the amino acid sequences of the histones are long-since determined.
But the structure of the DNA-histone complex, called chromatin, is not known. Does this strike you as odd? It certainly strikes me as odd. After all these years, with all the importance attached to gene engineering, even after completion of the "Genome Project", the structure of human chromatin remains unknown? Incredible!
Might it be that the histones simply can't be made to fit on a "double-helix" because the "double-helix" is not the structure of DNA in cells?
As we started to say above, the primary "business" of DNA is replication. In E.coli, a common bacterium, the replication cycle can be as short as 20 minutes. This is called the "log phase" of growth, because the number of bacteria in the culture doubles every 20 minutes, giving rise to a logarithmic increase in the bacterial count.
In the log phase, the daughter cells actually begin to split into two before they have finished separating from one another! Now, that's fast!
In 1963, an Australian researcher named John Cairns took a picture of the E. coli chromosome which found its way into every textbook of that period. The picture was called an "autoradiograph", meaning that E. coli bacteria were grown on a radioactive medium, and the cells were killed and split open over a photographic plate. That plate was put in a dark room for a few months, at the end of which the tiny quantity of radiation in the chromosomes gave rise to pictures. These pictures were examined under the electron microscope.
What was the structure of the E. coli chromosome which was revealed by these autoradiographs? It was circular. Subsequent to Cairns' great discovery, it was found that nearly all the DNA which can be isolated as a single intact chromosome is also circular. This includes all bacterial DNA which has been examined, all plasmid DNA from within eukaryotic cells, and most viral DNA.
Hmmm.... that's interesting. How does a circular chromosome replicate? The two strands of a circular double-helix are LOCKED TOGETHER. If you can't imagine that clearly, I'll show you a picture:

Figure A above shows a "classic" right-handed circular double-helix (B shows a "superhelix", which we won't have time to discuss just now).
If you can't clearly envision the linkage (called "topological linkage") between the two strands in Figure A, then please think of the links of a chain. Are the links of a chain locked together? Of course they are.
How is a chain made? You can make a short 2-link chain as follows: First you take a metal rod, and close it in a circle. Then you take a second rod, pass it through the circle, wrap it around once, and close it into a second circle. Are the two links of this chain locked together? Most assuredly.
To make DNA you take one single-stranded circle of DNA, pass a straight rod of DNA through it, and wrap it around, not once but thousands of times, then seal it shut. Are the two strands locked together? You'd best believe they are. See again Figure A above.
So, if the business of DNA is replication, and if the strands must separate at replication time, and if they are indeed locked together, then how do they separate when the cell divides? Good question!
Shortly after Cairns' great discovery of circular DNA, a major DNA structural symposium was held at Cold Spring Harbor, the "Mecca" of molecular biology. The subject of DNA replication, in the light of the recent discovery of circular DNA, was indeed featured. How prominently was it featured? Not very prominently. It was briefly mentioned in one paper, are largely glossed over.
According to the scientists at the Cold Spring Harbor Symposium, how did the strands of DNA get apart during replication? The answer they gave was a "swivel". What's a "swivel"? It's a hypothetical point, somewhat comparable to an automobile universal joint, around which the strands of DNA can untwist and re-twist during the process of unwinding of all those thousands of twists, and the re-winding of the twists back in the next generation.
The swivel was thought of as an enzyme which inserted itself into the chromosome, breaking it open, and "holding on" to the DNA strands as they whirled about, trying to unwind.
What's wrong with this picture? First of all, there was no "swivel" known! It was just a guess. What about the possibility that the hypothetical "swivel" was not needed, because DNA did not have the Watson-Crick structure in living systems, but rather some alternative un-twisted structure whose strands were free to separate at will? That possibility was not even mentioned. It had evidently been determined that, even at that early date, the "double-helix" was a "sacred cow", and could not be criticized. Therefore (or so the false logic went), since DNA had to be a double-helix, and since the strands had to separate during replication, there must have been a "swivel".
What had actually happened is quite clear in retrospect. Copernicus reduced man's arrogance by removing us from the center of the universe, and placing us in a measly little planet which revolved around one of billions of stars. The "double-helix", however, put us back, squarely in the center of the universe, since we would now "create life", and become "gods". What nonsense!
But the idea was compelling to many. It was compelling enough that, for the last 40 years, almost everyone who has criticized the "double-helix" has been miserably maltreated, and rejected from the mainstream of science.
It was quickly determined, after the announcement of the mythological "swivel", that the E. coli chromosome would have to be spinning at 6,000 rpm every minute of its life if it was to unwind all those twists every 20 minutes. This is too ludicrous for words. The E. coli chromosome is something like a millimeter long when stretched out, which is an immense length of DNA to stuff into a cell so small you can't even see it without a microscope. The very idea that all this DNA is whirling like an airplane propeller at 6,000 rpm is a joke.
"No problem". The helicists simply informed us that DNA only replicates a little bit at a time, so that only one small portion of the chromosome is actually spinning at any given moment. What evidence was there that DNA replicated this way? None. It was proposed, without evidence, to save the "swivel", which itself was proposed, without evidence, to save the "double helix". This is not science.
In 1976, an Australian researcher named Gordon Rodley, fed up with the obvious shenanigans going on with DNA, published an alternative structure, which is probably the correct one. In the Rodley structure, called the "side-by-side" structure (usually just called "SBS"), the two strands of DNA lie parallel to one another, undulating a bit to the right, then a bit to the left, but never making a whole twist. Here's a very over-simplified schematic of SBS, alongside another, less likely structure; both being examples of DNA structures in which the strands are not locked together:
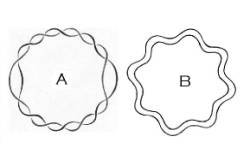
Figure A shows a totally hypothetical DNA structure which is "topologically equivalent" to the SBS structure, but in which the entire top half of the chromosome is a right-handed helix, and the entire bottom half is a left-handed helix. This model can be made instantly from two rubber bands by simply twisting them together. If you allow your rubber band model to "replicate", the two "strands" will separate as soon as you let go of them, because, topologically speaking, there were never really twisted together in the first place (every RH twist on top is canceled-out by a LH twist on the bottom). This structure, however, is highly unlikely to be found in nature.
Figure B shows the SBS structure, which is very likely to be found in nature, including in your own cells. What this drawing attempts to show is that the helix twists alternately a little to the right, then the left, etc., throughout the chromosome. As in Figure A, the net number of twists is actually 0. At replication time, the strands fall apart without having to untwist, because, topologically speaking, there really weren't any twists in the first place.
Rodley's structure (Figure B above) was published in the Proceedings of the National Academy of Science, one of the world's most rigorously peer-reviewed journals. How this got past the editors I'll never know, since Rodley remains, to this day, the only author whose work in this needlessly-controversial area has been accepted without a fight.
In 1978, Prof. Robert Chambers, the chairman of the biochemistry department at New York University School of Medicine, discovered by accident that the separated single-strands of the circular chromosome of the virus "phiX174" can spontaneously re-combine to give normal double-stranded circular DNA. This would be impossible if normal double-stranded circular DNA was a helix, because that would have required something like this:

In this drawing, a single-stranded circle is depicted as (1) being broken open, (2) wrapping itself around an intact circle, and (3) sealing itself shut again. The breaking open in step (1) is not a problem, because DNA can certainly break. The wrapping around the other strand in step (2) is not a problem because DNA is known to do that. But the sealing of the broken strand shut in step (3) is a BIG problem, because that requires a sealing enzyme, and no such enzyme was present in Prof. Chambers' single-stranded circular DNA preparation.
Is there another way to explain this data? Of course, but you have to be willing to believe that DNA is not a helix. It goes like this:

Here, the two circles are being shown simply coming together to form un-twisted double-stranded DNA. (Actually, it would be slightly twisted, but only as per the SBS structure two figures above this one).
Why has this simple concept been resisted so strongly? Because is was also discovered, not long after the discovery of circular DNA itself, that the two strands of circular DNA, when "melted", do not separate, which they ought to do if they are not locked together.
"Melting" doesn't mean literal melting, as when you heat butter in a frying pan. It means subjecting the DNA to conditions which promote strand separation. The first "melting" experiments were on fragmented linear DNA, and the procedure employed was simple boiling. At the temperature of boiling water, the double-stranded structure broke down, releasing single-stranded linear DNA. If the solutions were cooled slowly, the double-stranded form would, however, re-appear.
On the other hand, when circular DNA is boiled the strands do not separate. Moreover, at pH 13 (very, alkaline, like lye), where normal double-stranded DNA structure is impossible to maintain, the strands of circular DNA still do not separate! What's wrong?
I began working on this problem in the early 1970's. It took me years to solve the whole puzzle. In 1974 I submitted my first theoretical article on the subject to the Journal of Molecular Biology, which rudely rejected it. It took 28 years to get this article into a peer-reviewed scientific journal, but it was finally published a few months ago, in the Bulletin of Mathematical Biology (Biegeleisen, K. Topologically Non-linked Circular Duplex DNA, Bull Math Biol 64:589-609, May 2002).
The content of the article is a bit too long and drawn-out for the present discussion. Suffice it to say that the explanation for the well-documented fact that the strands of circular DNA do not separate under "melting" conditions is that the circularization of the DNA imparts novel and unexpected topological properties to the chromosome; properties which cause it to behave markedly differently from linear DNA under the same circumstances.
In particular, at high pH, where the single-strands of linear DNA fall apart, the single strands of circular DNA remain associated in a new, multi-strand, alkali-stable complex. The structure for this complex is similar to the structure originally proposed for all DNA by Linus Pauling, just months before the famous paper by Watson & Crick hit the press. Pauling, in his day, was considered to be the world's greatest living authority on the chemical bond, and the odds-makers had picked him to win the "race" to determine the structure of DNA. He considered a triple or quadruple helix with the phosphate groups pointing inward to be the most logical structure for DNA, based on the chemistry only (i.e., without reference to genetically-specific base-pairing, which had not yet been discovered).
When Watson & Crick published the "double helix", it looked as if Pauling had "lost the race". Now, however, almost exactly 50 years later, we can see that maybe he didn't lost it entirely.
If circular duplex DNA really has the SBS structure, or one similar to it, then it ought to be possible to separate the component single-stranded circles, without breaking either one open. This was not accomplished for many years.
Proof that the strands of circular DNA can indeed be separated was finally published in 1996, by a brilliant researcher named Tai Te Wu, with the help of his son (Wu R. and T.T. Wu, 1996. A novel intact circular dsDNA supercoil. Bull Math Biol, 58(6):1171-1185).
Wu's innovation was to isolate double-stranded chromosomal DNA from a plasmid under conditions where a great deal of a substance called "RNA" was bound to only one of the two strands of the DNA chromosome. The bound RNA changed the physical characteristics of the strand it was bound to, so that Wu was able to separate the two chromosomal strands by prolonged gel electrophoresis (a procedure in which DNA's of different physical characteristics can be separated into distinct bands in a jello-like substance to which an electric current has been applied). Wu repeated the work with a second plasmid which had the two strands marked by the intentional insertion of marker sequences by genetic engineering, and proved, unequivocally, that the two bands in the gel were indeed the two single-strands of the original plasmid chromosome, respectively.
The conclusion is that DNA is not a helix, and it's really been apparent all along to many people. The failure of the two strands of circular DNA to separate under "melting" conditions made it possible for the scientific community to evade this obvious conclusion for many years, but the days of the "double-helix" as the presumed structure of DNA in living system are, in all probability, numbered.
Ken Biegeleisen
Running head: Topologically non-linked DNA
Author's address:
Ken Biegeleisen, M.D., Ph.D.
133 East 73rd Street
New York, N.Y. 10021
U.S.A.
Tel (212) 717-4422
(no fax)
Email: biegel@VeinDoctorNY.com
Method of submission: Electronic. (Original figures available if necessary).
Original materials: If figures are submitted on paper, I will need the originals back.
The discovery of circular DNA, over 30 years ago, introduced an element of uneasiness in what had been, up to that point, the almost picture-perfect story of the elucidation of the molecular biology of heredity. If DNA indeed has the Watson-Crick right-handed helical secondary structure, then in circular DNA, thousands, or perhaps even millions of twists must be removed in each generation, and re-wound in the next generation.
Although enzyme systems adequate for this task have long since been found and characterized, there have nevertheless arisen a number of proposals for alternative DNA structures in which the strands are topologically non-linked, so that they might separate during replication without having to be unwound. These structures have generally been put forth as theory only, and have been largely unaccompanied by experimental evidence to support their applicability to native DNA from living systems.
Recently, however, a report has emerged suggesting that it might be possible to separate, intact, the individual single-stranded circular half-chromosomes which constitute the double-stranded circular chromosomes of certain plasmids. This would not be possible unless the chromosomes had one of the alternative, topologically non-linked structures.
It is widely believed that after a half-century of worldwide DNA research, any significant change to the Watson-Crick structure is unlikely to stand up to scrutiny. Nevertheless, the present author has found that in many instances in which the behavior of circular duplex DNA is considered to be explicable only in terms of the topologically-linked helical model, it is also possible to explain that same behavior in terms of a topologically non-linked model. It is necessary, in these instances, to make certain logical assumptions which cannot be conclusively proven at the present time.
The author herein offers an example of one such instance, namely an examination of the behavior of circular duplex DNA in an alkaline titration experiment, where conformational changes in DNA are deduced from changes in its buoyant density at pH's between 7 and 14. These data have been explained in terms of topological linkage between the DNA strands, but they can also be explained without invoking any such topological linkage, provided that the above-mentioned logical assumptions can be accepted.
The principles which emerge from this are applicable to other settings in which knowledge of the topology of DNA is critical to the understanding of observed phenomena.
Wu & Wu (1996) reported, in these pages, the separation of the intact circular single strands comprising the circular duplex chromosomes of 2 different plasmids. These authors have either made a grand error, or else have discovered a system within which DNA does not have the right-handed helical structure, the strands of which, when circularized, would not be separable unless at least one of them was broken open.
These authors believe that circular DNA is not, in general, topologically helical, but rather has a structure not unlike those proposed by Rodley et al (1976) and Sasisekharan & Pattabiraman (1978), in which the two individual single-stranded circular half-chromosomes twist about each other alternately to the right and left, giving rise ultimately to a structure whose strands are topologically non-linked. Since the structures proposed by each of the above authors differ somewhat, I shall refer to them collectively by the initials TN DNA, for topologically non-linked DNA (Fig. 1).
It has been known for decades that the individual single strands which comprise the double-stranded chromosomes of most species of circular DNA do not separate under conditions commonly observed to cause strand separation in either linear DNA, or in circular duplex DNA which has had one or both strands nicked (Vinograd et al., 1965) (Rush & Warner, 1970). Although the secondary structure of fully-intact circular duplex DNA is indeed disrupted by denaturation, the strands nevertheless remain associated with each other, as if they were physically locked together. It is not at all surprising that this observation has discouraged many from seriously considering any proposals for alternative structures for DNA in which the strands are not topologically linked.
In order to attain the separation of the intact individual circular single strands from duplex circular DNA, Wu & Wu (1996) employed the ingenious subterfuge of growing plasmids in stationary phase cells, where there is little DNA replication (and therefore few replicative intermediates to confuse things), but ongoing transcription. Since the RNA is transcribed only from the sense strand of DNA, and since DNA-RNA hybrids on gel electrophoresis have been found to be more stable than DNA-DNA hybrids (Casey & Davidson, 1977), Wu & Wu deduced that the electrophoretic mobility of the sense and "nonsense" strands were not the same, and that they could be separated by gel electrophoresis under the proper conditions. The data they have presented appears to confirm their prediction, at least in the system they have employed.
It is not the purpose of this paper to take any position on the structure of DNA in general, which will ultimately be determined by research and observation. Rather, this manuscript will focus on a single hypothetical question. We shall assume that Wu & Wu (1996) are correct, and that circular DNA, at least in the systems they have reported on, has a TN structure. The question is this: If any DNA has the structure Wu & Wu propose, how can we account for the failure of the strands to separate under more usual conditions of denaturation, such as alkali denaturation, where decades of observations have confirmed repeatedly that the strands do not separate?
When one considers the question carefully, one's thoughts are compelled in particular directions by the body of data which exists concerning the behavior of circular DNA in various settings. We find that we must make certain assumptions. Each of these assumptions can be readily challenged, and we cannot assert that they are correct, but only that they find some support in existing evidence. I therefore present them as theory, in the spirit of the statement made by Crick et al. (1979): "DNA is such an important molecule that it is almost impossible to learn too much about it".
If DNA has any of the TN structures which have been proposed to date, then it would be perfectly logical to expect that the strands would separate upon denaturation. They do not. I shall first present, without comment, the assumptions necessary to explain this. Then I will review some experimental evidence suggesting that these assumptions are at least possible. Finally I will show how they can be applied to answer the question we have raised.
1. It must be assumed that DNA generally has the propensity to exist as either a right-handed helix or a left-handed helix, depending upon the prevailing conditions.
2. It must be assumed that conditions favoring denaturation will generally bring about a transition from the right-handed to the left-handed secondary configuration. For brevity, I shall refer to this as an RÆL transition.
3. It must be assumed that the duplex product resulting from denaturation of circular DNA has an ordered structure requiring the participation of both strands, by means of which we may account for the failure of the strands to separate even though they are not topologically locked together.
Left-handed DNA has been known for many years (Pohl & Jovin, 1972) (Ikehara et al., 1972) (Pohl 1976) (Wang et al., 1979) (Mitsui et al., 1970) (Nordheim et al., 1981). It was first observed in synthetic copolymers. It is not considered to be the structure of purified DNA in solution for any DNA obtained from natural sources, so that the presumption that DNA from natural sources can exist in the left-handed configuration is unproven. Nevertheless, we need to make this presumption for the sake of the argument which follows later.
There has existed evidence, for many years, that ordinary DNA might undergo an RÆL transition under conditions which promote unwinding. Travers et al. (1970) found that the optical rotatory dispersion (ORD) spectrum of purified DNA inverted in aqueous methanol solutions. They suggested that this was best explained as an RÆL transition, although there are other possible explanations. Similar inversions were demonstrated in the circular dichroism (CD) spectra of DNA at high salt concentration (Zimmer & Luck, 1974), and following complexing with mitomycin C (Mercado & Tomasz, 1977).
In the case of certain synthetic polynucleotides, the nature of these spectral inversions was further studied by x-ray crystallography (Mitsui et al., 1970) (Wang et al., 1979). These authors concluded that the polynucleotides were left-handed.
Since methanol, high salt, and Mitomycin C have little in common chemically, I will propose that anything which unwinds DNA may, under the right conditions, bring about an RÆL transition. This is obviously a sweeping assumption, but it is necessary for the argument we shall present.
The assumption is not without logical support. Such can be found in the work of Wang et al. (1979) on the pitch of the left-handed helical DNA fragment d(CpGpCpGpCpG); whose structure they referred to as ìZ-DNAî. This helix has a rise per residue of 3.7 angstroms, which is considerably larger than the 3.4 angstrom spacing between the bases of the ìnormalî right-handed DNA helix. With 12 residues per helical turn in Z-DNA (compared with 10 for right-handed DNA), it has a pitch of 45 angstroms (compared with 34 angstroms for right-handed DNA).
In other words, Z-DNA appears to be a more loosely wound helix than right-handed DNA. If this turns out to be generally true of left-handed DNA, then it follows logically that any substance which tends to unwind right-handed DNA might, if added in increasing quantity, cause the left-handed form of DNA to eventually become the favored form.
As for our final assumption, the existence of a non-topologically-linked ordered duplex structure for denatured circular DNA, there is little which can be said with certainty. In days gone by I can still recall hearing molecular biologists describing the structure of denatured circular DNA as "a tangled mess", as if the single strands had no interaction with one another at all, other than the physical constraint imposed by the presumed topological linkage. Early attempts to renature it failed, and the term "irreversibly denatured" was employed at first. The studies of Robert Warner (Strider and Warner, 1971; Strider, 1971; Strider et al 1981), however, showed that denatured circular DNA can indeed be renatured, but only under very narrow conditions of pH, temperature and ionic strength (Fig. 2). These data certainly do not suggest anything "random" about the structure of denatured circular DNA, but rather a highly ordered structure; one which responds in a very exact way to even minute changes in the environment. But whether that structure is topologically linked or not cannot be determined from these data.
I shall adhere to the original Roman Numeral system for describing circular DNA and its cleavage products.
Form I: The replicative form of most small circular DNA. Form I is a covalently closed, circular duplex chromosome. The electron micrographic appearance of this species is rarely an open circle, however, because it is usually isolated in a form bearing tertiary superhelical twists (Fig. 3).
Form II: The duplex product resulting from the introduction of one or more single-stranded nicks into Form I. Even a single nick into either strand of Form I causes the tertiary superhelical twists to unwind. The chromosome is then relaxed, appearing in electron micrographs as an open circle.
Form III: The linear duplex product arising from the full duplex cleavage of Form I.
Form IV: The product resulting from the alkali denaturation of Form I. It is an extremely dense duplex of uncertain structure.
We are now in a position to apply the above assumptions to explain the failure of the strands of Form I to separate during ordinary denaturation, without invoking topological linkage. Because it was hoped, at one time, that alkali denaturation might prove to be a useful tool for purification of Form I DNA on a large scale, it was studied very closely. We shall therefore take advantage of those abundant data to examine our model.
Fig. 4 shows a typical pH vs. sedimentation coefficient curve for a small circular DNA (that of the bacteriophage fX174). This curve has been observed for a variety of small circular DNA's (Rush & Warner, 1970) (Vinograd et al., 1965), and it seems to be characteristic of naturally-occurring small circular DNA's of average base composition. It has a number of important aspects, each of which we shall presently have need to consider. For the moment, however, it is sufficient to note that at high pH, the two strands of double-stranded nicked circular DNA (i.e., Form II) separate into single-stranded circles and linear forms, whereas the strands of intact duplex circular DNA (Form I) do not. The latter molecule, rather, remains double-stranded even at pH's greatly in excess of those required for denaturation. This denatured duplex product is the form known as ìForm IVî.
The traditional explanation for these observations depends upon the presumption that the strands of Form I DNA are topologically linked. As one can readily persuade oneself by working with circular chromosome models constructed from bits of string, two circular structures which are twisted together before being sealed shut into a circle are, in fact, physically locked together. This is referred to as ìtopological linkageî, and is treated as a quantitative property of circular DNA, having its own numerical parameter alpha (a), the ìtopological winding numberî (Glaubiger & Hearst, 1967), (or, alternatively, ìLkî, the ìlinking numberî)(Crick et al., 1979).
Fig. 3 depicts an all-right-handed Watson-Crick circular double helix. The reader who cannot clearly envision the topological linkage between the two strands is strongly urged to make a model from two pieces of string. Without a clear picture of topological linkage, the remaining arguments to be presented will be impossible to grasp.
I will give a brief description of the traditional explanation for the data in Fig. 4, then move on to the explanation in terms of the TN model. The traditional explanation is as follows: At neutral pH, the sedimentation coefficient of Form I is greater than that of Form II because Form I is superhelical. The superhelices are presumed to be the result of the chromosome having been sealed shut in an underwound state, meaning that although it has essentially the Watson-Crick right-handed helical structure, it has, for some reason, a deficiency of right-handed turns. These are compensated for, one-for-one, by the appearance of right-handed superhelical turns (for reasons which will be explained presently). As the pH increases past 11.5, the DNA begins to denature ("melt"), and the chromosome starts to unwind, with the superhelical turns coming out first. As they do so, the chromosome proceeds to relax, until, at a pH just below 12, it becomes an open circle, with the same sedimentation coefficient as Form II. At higher pH's, the secondary structure "tries" to unwind, but cannot, due to the presumed topological linkage. Therefore, it assumes an ever-increasing number of left-handed superhelical turns (each of which causes the unwinding of a right-handed secondary helical turn; see below), until, at some pH in the range 12-13, it reaches the point of "irreversible" denaturation into Form IV. The sedimentation coefficient of Form IV is very high, indicating a dense, compact structure. If solutions of Form IV are neutralized (the dashed line at the top of Fig. 4), they become somewhat less dense, but they remain much denser than the Form I from which they arose.
In order to proceed, we will need a way to visualize TN DNA. Fig. 1A shows one such way. In this picture, TN DNA is depicted as consisting of a single long right-handed segment and a single equally long left-handed segment.
This "model" emphasizes a critically important topological feature of TN DNA, namely that it is topologically 50% right-handed and 50% left-handed. But it certainly seems unlikely that such a structure will be observed in nature.
A more plausible structure would be the "side-by-side" structure of Rodley et al. (1976), who proposed no complete helical turns at all, but rather a quasi-helical structure which alternately winds a bit to the left, then to the right, without ever making a full helical turn (Fig. 1B).
It should be specifically noted that the Rodley side-by-side structure is topologically 50% right-handed and 50% left-handed, only the length of each of the many alternating helical regions is less than one full turn, so that the strands, when constrained to lie in a plane, never cross one another. DNA with such a structure could replicate without the requirement of a mechanism for the unwinding of secondary twists.
Since x-ray crystallographic studies have shown that purified crystals of linear DNA molecules from natural sources are right-handed helices, we may assume that the common DNA's will generally ìpreferî to be right-handed, insofar as it is topologically possible for them to be so.
But in TN DNA, it is not topologically possible. Rather, exactly and precisely half the molecule must be left-handed, if the other half is to be right-handed (as can be seen in Fig. 1, or more readily by constructing string models). The TN chromosome is therefore under strain at neutral pH, since it "wants" to unwind its left-handed helical portions, but cannot.
In solutions of TN DNA, however, it would be possible for the individual molecules to form superhelices (see Fig. 5). The relationship between right-handed helical turns and superhelical turns in Form I DNA is fixed by the laws of topology . The introduction of a single 180 tertiary, or superhelical turn into circular duplex DNA causes the unwinding of a single 360 secondary helical turn of the opposite sense (Glaubiger & Hearst, 1967). In Figs. 5A & 5B, for example, the introduction of two such left-handed tertiary superhelical turns causes the unwinding of two right-handed secondary helical turns. Again, the reader who cannot accept these topological facts from the figure alone is urged to verify them by constructing a string model.
To state these facts anthropomorphically, under conditions in which DNA ìwantsî to unwind its secondary left-handed helical turns, it can accomplish this by assuming superhelical turns in the opposite sense.
Once these topological facts are appreciated, it can immediately be seen why TN DNA must be superhelical at physiological pH: It ìprefersî the right-handed helical conformation, but must, by reason of topological restraint, be at all times exactly and precisely 50% left-handed, when constrained to lie in a plane. In order to maximize its right-handedness, it therefore refuses to lie in a plane. Rather, it takes on as many right-handed superhelical turns as possible, each of which causes the unwinding of ìunwantedî left-handed secondary helical turns.
We have therefore provided an explanation for the known fact that native Form I DNA is isolated as a superhelix (Vinograd et al., 1968) (Glaubiger & Hearst, 1967) (Shure & Vinograd, 1976), without presupposing any topological linkages between the strands. We have also correctly predicted the direction of Form I superhelical winding, which is, in fact, known to be right handed.
Therefore, the well-documented fact that Form I DNA is superhelical, and hence more dense, with a higher sedimentation coefficient (s) than its nicked Form II "cousin" (Fig. 4), is not necessarily incompatible with the theory that Form I has the TN structure.
Let us now return to Fig. 4 and consider the next step in denaturation of Form I DNA, which occurs between pH 11.5-12.0. In this region of the curve s decreases to a value similar to that of Form II nicked, relaxed DNA. As in ìtraditionalî theory, we shall presume that this occurs because of the unwinding of superhelical turns, yielding a less compact molecule. According to our theory of TN DNA, this is consistent with the expected behavior of the molecule, since we have assumed that conditions tending toward denaturation favor an RÆL helical transition.
In other words, as the pH increases, right-handed helical DNA starts to unwind, causing the left-handed helical form to become increasingly favorable until, at some pH between 11.5 and 12.0, the right-handed and left-handed forms become energetically equal. At that point, Form I DNA no longer ìcaresî which way it twists. So it relaxes, the superhelices unwind, and the molecule appears indistinguishable at that moment from Form II relaxed DNA.
We have therefore again accounted for the behavior of Form I DNA in Fig. 4 without assuming any topological linkages between the strands.
We now arrive at the most critical part of the curve. As the pH approaches 12, s begins to increase for both Forms I and II. For Form I this is readily explained. But why should this occur with Form II? The ìtraditionalî theory of DNA as a right-handed helix offers no explanation. But the theory of TN DNA, which incorporates the observation of the tendency of DNA to convert to the left-handed helical form under conditions tending toward denaturation, states simply that for both Forms, I and II, the DNA begins to express a ìwishî to be left-handed, which translates into left-handed superhelix formation (since left-handed superhelical turns unwind right-handed secondary helical turns - see Figs. 5A & 5B).
Next, at pH's above 12, Form II splits into separate strands of linear and circular single-stranded DNA, and its sedimentation coefficient, s, returns to that of relaxed DNA (Fig. 4).
But Form I DNA behaves differently. No denaturation into single-strands is observed. Why not?
It is possible to identify at least two reasons why Forms I & II DNA might behave differently at pH 12. In order to understand them, we must do a "thought experiment", mentally shrinking ourselves to the size of a denaturing TN DNA molecule, and asking the question "What would we actually see?".
The first thing we would see is that Form I, topologically speaking, is 50% left-handed at all times. Mathematically, this may be expressed by the statement
...alpha being the ìtopological winding numberî mentioned earlier. This means that if, in our minds, we constrain the molecule to lie in a plane (without superhelical twists), the number of right-handed and left-handed helical turns must always be seen to be exactly the same, and that the net number of secondary helical turns in the entire molecule must always add up to zero.
This means that there is a topological constraint on the molecule, such that every right-handed helical turn which is to be unwound must be accompanied by the compensatory unwinding of a left-handed helical turn also (or, if the molecule is not constrained to lie in a plane, the compensatory winding in of a left-handed superhelical turn).
These are not biological facts, but mathematical truths, fixed by the laws of topology. Again, see Fig. 5, or simply hold two rubber bands together and twist them.
It is entirely different with Form II, which has no topological constraints. Thus, our theory states that at pH 12, Form II, after briefly taking on some left-handed supertwists (note again the increase in s in Fig. 4 at pH 12), ìdecidesî to convert itself quantitatively into an all-left-handed helix, and there is no topological barrier to it doing so. So it tries to twist to the left. But what happens when it tries to do so?
One can readily envision Form II, at this pH, spinning rapidly about its long axis, like an electric drill bit. This process, occurring at high pH where base-pairing interactions are vastly weakened, must surely be a disruptive process; one which encourages denaturation by the sheer centrifugal force of the cooperative RÆL transition. In fact, Form II, at this pH, does indeed denature, yielding single-strands.
I am suggesting, of necessity, that Form II denatures at a pH where base-pairing, although weakened, is not gone entirely, and that the event which triggers denaturation is therefore not the loss of base-pairing, but rather the rapid rotation of Form II about its long axis, as it tries to convert itself into a left-handed helix.
In Form I DNA, on the other hand, the above-mentioned topological constraint bars the possibility of a wholesale ìdrill-bit-likeî RÆL conversion. Why? Because, as we have noted (or as may be seen by twisting-untwisting a pair of rubber bands), every right-handed helical turn which is removed must be accompanied by the removal of a left-handed helical turn. But the left-handed turns do not "want" to be removed at this pH; on the contrary, they want to increase. Thus, they will resist being unwound. Clearly, therefore, there will be no rapid RÆL transition in Form I at pH 12.
The "desire" to become left-handed at high pH, however, can be satisfied, at least in part, by superhelix formation. Thus, the molecule will twist itself into a left-handed superhelix, which will bring about the removal of "unwanted" right-handed secondary turns without the necessity of simultaneously removing the now-desirable left-handed secondary turns.
Therefore, at pH's around 12, the ultimate effect of increasing the pH of solutions of Form II DNA is rapid and abrupt cooperative unwinding leading to strand separation, but the effect for Form I DNA is merely the gradual formation of ever-increasing numbers of left-handed superhelical turns.
Thus, without pre-supposing topological linkages between the strands of Form I, we have explained the alkali denaturation curve of Form I up to pH's of around 12. We still have not, however, explained why the strands of Form I donít separate at pHs above 12.
In our "thought experiment", we may make the following further observation about Form I DNA: it has no free end.
Denaturation of linear DNA has always been thought of as being initiated at a free end. As Fig. 6 shows (in highly schematic form), it will, as a first approximation, require fully twice as much disruptive energy to initiate denaturation in a covalently-closed-circular molecule as in a nicked one which has a free end.
The degree of protection against strand separation in Form I can be roughly estimated. In Fig. 6 we have represented the divisive forces of denaturation as if they were the forces of a miniature weight-lifter lifting physical weights. It may readily be seen that it will, as a first approximation, require twice as much force to initiate denaturation in an intact duplex circle with no free end. But in reality, it is not the lifting of weights, of course, but the hydroxide ion concentration which constitutes the divisive force. Therefore, a doubling of this force may, as a first approximation, be thought of as a doubling of the hydroxide ion concentration, corresponding to a pH increment of about 0.3.
As a matter of fact, there is a shoulder on the pH vs. s curve of Form I at about 0.3 pH units above the point where its Form II ìcousinî dissociates into single-strands. This shoulder is labeled ìcî in Fig. 4. In our TN theory, this corresponds to the point at which the above-mentioned cooperative protection against denaturation expires, and TN DNA finally denatures, converting from superhelical Form I into the mysterious ìForm IVî. All theory aside, the fact that this is indeed the true point of irreversible conversion to Form IV has been confirmed by the painstaking work of Strider et al (1981).
Why then does TN Form I not denature into single-stranded DNA? The only possible answer is that the formation of a tightly-wound superhelix brings about a condition conducive to the formation of a new tertiary structure; one which is stable at high pH. What might it be? Obviously, it cannot be said with certainty. Based upon what is known about multi-chain DNA structure, both theoretically and empirically, two broad categories of structure suggest themselves: multi-chain structures with the phosphate groups on the inside, and multi-chain base-paired structures.
What is the structure of Form IV? The question, in this direct form, cannot be answered.
If we alter the question slightly, however, we can do better. Why might the strands of denaturing Form I DNA be predisposed to forming multi-chain structures, whereas in other forms of DNA the strands fall apart?
The question, in this form, can be answered. Getting back to our "thought experiment", we note that in Form I DNA at pHs above 12, there are four DNA strands lying close together (Fig. 7A-C). The molecule is becoming increasingly superhelical at these pH's; much more so than is ever seen at physiological pH, where the superhelix is relatively loosely wound. According to what criterion can we say this? According to the criterion that at pHs around 12.3, s for Form I is nearly twice its value at pH 7 (see Fig. 4), which suggests that the superhelix surely was loosely wound at physiological pH, and has now become dramatically more tightly wound at pH 12.3. Consider what this means: the two sides of the double-helix have been progressively twisted together, with water being "squeezed" out of the core, so that there are literally four DNA strands lying closely juxtaposed (Figs. 7A-C). This is a circumstance which never occurs with nicked DNA (it begins just below pH 12, but is interrupted by total denaturation and strand separation).
It is not at all difficult to imagine this forced proximity of four DNA strands giving rise to structures which might not form spontaneously in other settings. But what might these structures be? We shall consider two.
In 1953, just before publication of the Watson-Crick-Wilkins-Franklin model of right-handed double-helical DNA, Linus Pauling (Pauling and Corey, 1953) published a paper suggesting a three- or four-stranded structure held together by phosphate salt bridges. Pauling, a double Nobel-laureate, was, according to James Watsonís popular book The Double Helix, the worldís leading authority on the chemical bond, and the scientist considered most likely to win the race to discover the structure of DNA.
Lacking knowledge about genetically-specific base-pairing, Pauling found that the best way to construct a helix from DNA, from the purely chemical point of view, was to put the phosphate groups in the inside. His structure, a three-stranded helix with the bases on the outside, had the phosphates on the inside, close-packed into tetrahedral formations, giving rise to a helix with the required pitch of 34 angstroms. He chose a three-stranded structure because it was more readily made to accommodate itself to the existing x-ray crystallographs of DNA, but it was pointed out that it was easier still to pack the phosphates into a four-stranded structure (Pauling and Corey, 1953).
It has been known for many years that there actually are examples in nature of viral DNA helical chromosomes with the phosphate groups on the inside. Loren Day, of the Public Health Research Institute of New York City has identified two such viruses (Day et al, 1979) (Liu & Day, 1994).
One possible structure for Form IV, therefore, would be a four-stranded helix stabilized by phosphate salt bridges (Fig. 7D), not unlike the structure originally proposed by Pauling as the structure for all DNA.
More recently, Alexander Rich and his co-workers have identified four-stranded DNA structures which are base-paired (Kang et al, 1994) (Kang et al, 1995). These are described as consisting of two pairs of duplexes, each of which consist of two parallel strands held together by atypical hydrogen bonds (cytosine-protonated cytosine, or C-C+ base pairs). The two base-paired duplexes are then intercalated into each other in opposite orientations.
These structures were identified in cytosine-rich DNA's, namely the copolymer d(C3T) and the telomeric cytosine-rich repeating sequence d(TAACCC).
If DNA has a TN structure, then in the four-stranded configuration which exists just before the complete alkali denaturation of Form I (Fig. 7C), the possibility exists for two pairs of parallel strands to intercalate into each other in opposite orientations. Whether atypical base-pairing, however, will be seen in more typical species of DNA which are not necessarily cytosine-rich, I cannot say.
In any event, since the existence of any significant base-pairing at high pH seems unlikely, we would have to presume that such structures at high pH were stabilized solely by base stacking interactions.
All these things considered, I favor the Pauling model of DNA for the structure of Form IV.
Whatever the structure of Form IV, it is not based on genetically-specific hydrogen bonding, so the complementary bases do not remain in register.
I would suggest, based upon what little we know about the circumstances of its formation (pretty much all of which is shown in Fig. 7), that Form IV is a quadruple helix of some type. If so, then we can say also that the tightness of the quadruple helical winding can be varied by varying the pH (see the upper curve in Fig. 4), but that it remains a non-base-paired quadruple structure under all circumstances. All, that is, except those narrow sets of circumstances described by Warner and his coworkers (Fig. 2), under which renaturation to Form I takes place.
The explanation for the behavior of Form I DNA in terms of the theory of topologically non-linked structure may be reduced to five basic principles:
1. The individual strands of circular duplex DNA in nature are presumed to be topologically non-linked, giving rise to a duplex chromosome having a structure such as has been proposed by several authors. Topologically, these structures are 50% right-handed and 50% left-handed, arranged as short, regularly alternating helical regions each less than one full turn in length.
2. Purified solutions of topologically unrestrained DNA (i.e., linear DNA or Form II nicked DNA) are presumed to have the "traditional" right-handed helical structure at neutral pH, and the left-handed helical structure under conditions which promote unwinding.
3. At neutral pH, purified solutions of Form I TN DNA have the right-handed superhelical tertiary structure, because this structure maximizes the right-handedness of the secondary winding. This it does by converting "unwanted" left-handed secondary helical turns into right-handed tertiary superhelical turns.
4. At higher pH (i.e. about pH 12), purified solutions of Form I DNA have the left-handed superhelical tertiary structure, because this structure maximizes the left-handedness of the secondary winding. This it does by converting "unwanted" right-handed secondary helical turns into left-handed tertiary superhelical turns.
5. At pH's above 12.3, the left-handed superhelical winding is so tight that four DNA strands are forced into perfect alignment, overcoming the activation energy for the formation of a new structure: a four-stranded helix, most likely one with the phosphate groups in the core, stabilized by salt bridges.
These five principles may be employed to explain other topological phenomena reported for Form I DNA, including the electron microscopic appearance of replicative intermediates (Jaenisch et al., 1971; Sebring et al., 1971), the appearance and distribution of discreet bands in topoisomerase experiments (Crick et al., 1979), and the observation of greater stability of DNA-RNA hybrids than DNA-DNA hybrids during gel electrophoresis (Casey & Davidson, 1977). Space limitations preclude detailed discussions of these phenomena.
Stettler et al. (1979) set out to disprove all TN hypotheses by showing that the reannealing of separated single-stranded circles of complementary DNA did not produce Form I, as would have been required by any TN theory. In the place of Form I, these authors reported the appearance of an anomalous duplex structure they dubbed "Form V". Their report ought not to be accepted uncritically, since their experiment was essentially uncontrolled (the control experiment employed a different DNA, in a different solvent, at a different temperature than the real experiment) (see also Fig. 8 in the present manuscript). Furthermore, although most of the Form V structure was alleged to be in the double-helical base-paired conformation, the thermal denaturation profile showed no cooperativity at all (see Stettler et al.ís Fig. 13).
They had a "competitor" in this work, Dr. Robert W. Chambers, at that time acting chairman of the Biochemistry Department at the New York University School of Medicine, where I was a graduate student. Prof. Chambers had also set out to prove that one cannot make Form I from complementary single-stranded circles, but wound up proving just the opposite! After becoming aware of the publication of the Stettler paper, Chambers retired his painstakingly-isolated preparation of complementary single-stranded circular DNA to the refrigerator. Three months later, a significant portion of it had turned into Form I. Chambers, a staunch "traditionalist", was unwilling to challenge the Watson-Crick theory, and, perhaps because he was unable to provide a satisfactory explanation for his discovery in terms of "traditional" theory, he chose not to publish it (R.W. Chambers, personal communication, 1978).
I believe that the best way to formulate a strategy for definitively testing the TN hypothesis is to start by noting that Form IV, in the final analysis, is a type of separated complementary single-stranded circular DNA. It may therefore be that the problem of getting the single strands of Form IV to re-anneal may not be so different than the analogous re-annealing problem when the starting material is separated complementary single-stranded circles floating freely in solution.
Under the conditions of optimum temperature and pH shown in Fig. 2, denatured circular DNA is induced to enter into a conformation from which it readily passes into Form I.
If, as the TN theory states, the individual strands of Form IV are topologically non-linked, then separated complementary single-stranded circles, floating freely in solution, ought to associate with one another in much the same way that aggregated complementary single strands associate in Form IV, under the conditions of temperature-pH optima depicted in Fig. 2.
Therefore, incubation of separated complementary single-stranded circles under these conditions-and these conditions only-ought to give rise rapidly and quantitatively to Form I DNA. Incubation under any other conditions should be avoided, since, as Fig. 2 suggests, small alterations of pH and temperature might have profound effects on the outcome of such an experiment.
Fig. 8 is a plot of the temperature-pH optima from Fig. 2. Incubation of separated complementary single-stranded circles under any of these conditions ought to yield rapid and quantitative conversion of the single-stranded circles back to normal Form I duplex circular DNA.
In any laboratory possessing complementary single-stranded circular DNA, this experiment can be done in a single day.
Another way to test the TN hypotheses would be to take advantage of the data in Fig. 8 to try to separate the strands of Form I by boiling. Under physiological conditions of pH and ionic strength, Form I does not denature when boiled. But if the data in Fig. 8 were extrapolated to the temperature of boiling water, Form I might behave as linear DNA does, and separate into single strands. This experiment is not "clean", however, because boiling causes scission of DNA strands. Also, relative to the stringent temperature-pH requirements made evident in Fig. 2, it may be that the data in Fig. 8 are not precise enough to determine the conditions of this experiment with certainty.
REFERENCES
Cairns, J. (1963). The chromosome of Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 28, 43-46.
Casey, J. and N. Davidson (1977). Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucl. Acids Res. 4, 1539-1552.
Crick, F.H.C., J.C. Wang and W.R. Bauer (1979). Is DNA really a double helix? J. Mol. Biol. 129, 449-461.
Day, L.A., R.L. Wiseman and C.J. Marzec (1979). Structure models for DNA in filamentous viruses with phosphates near the center. Nucl. Acids Res. 7(6), 1393-1403.
Glaubiger, D. and J.E. Hearst (1967). Effect of superhelical structure on the secondary structure of DNA rings. Biopolymers 5, 691-696.
Ikehara, M., S. Uesugi and J. Yano (1972). Left-handed helical polynucleotides with d-sugar phosphodiester backbones. Nature New Biol., 240, 16-17.
Jaenisch, R., A. Mayer and A. Levine (1971). Replicating SV40 molecules containing closed circular template DNA strands. Nature New Biol. 233, 72-75.
Kang, C., I. Berger, C. Lockshin, R. Ratliff, R. Moyzis and A. Rich (1994). Crystal structure of intercalated four-stranded d(C3T) at 1.4 angstrom resolution. Proc. Natl. Acad. Sci. USA, 91, 11636-11640.
Kang, C., I. Berger, C. Lockshin, R. Ratliff, R. Moyzis and A. Rich (1995). Stable loop in the crystal structure of the intercalated four-stranded cytosine-rich metazoan telomere. Proc. Natl. Acad. Sci. USA, 92, 3874-3878.
Kasamatsu, H. and M. Wu (1976). Structure of a nicked DNA-protein complex isolated from simian virus 40: Covalent attachment of the protein to DNA and nick specificity. Proc. Natl. Acad. Sci. USA. 73, 1945-1949.
Liu, D.J. and L.A. Day (1994). Pf1 virus structure: helical coat protein and DNA with paraxial phosphates. Science 265, 671-674.
Mercado, C.M. and M. Tomasz (1977). Circular dichroism of mitomycin-DNA complexes. Evidence for a conformational change in DNA. Biochemistry 16, 2040-2046.
Mitsui, Y., R. Langridge, B.E. Shortle, C.R. Cantor, R.C. Grant, M. Kodama, and R.D. Wells (1970). Physical and enzymatic studies on poly d(I-C).poly d(I-C), an unusual double-helical DNA. Nature 228, 1166-1169.
Nordheim, A., M.L. Pardue, E.M. Lafer, A. Mller, B.D. Stollar and A. Rich (1981). Antibodies to left-handed Z-DNA bind to interband regions of Drosophila polytene chromosomes. Nature 294, 417-422.
Pauling, L. and R.B. Corey (1953). A proposed structure for the nucleic acids. Proc. Natl. Acad. Sci. USA. 39, 84-97.
Pohl, F.M. and T.M. Jovin (1972). Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly(dG-dC). J. Mol. Biol. 67, 375-396.
Pohl, F.M. (1976). Polymorphism of a synthetic DNA in solution. Nature 260, 365-366.
Pohl, W.F. and G.W. Roberts (1978). Topological Considerations in the Theory of Replication of DNA. J. Math. Biol. 6, 383-402.
Pouwels, P.H., J. Van Rotterdam and J.A. Cohen (1969). Structure of the replicative form of bacteriophage fX174. VII. Renaturation of denatured double-stranded fX DNA. J. Mol. Biol. 40, 379-390.
Rodley, G.A., R.S. Scobie, R.H.T. Bates and R.M. Lewitt (1976). A possible conformation for double-stranded polynucleotides. Proc. Natl. Acad. Sci., USA. 73, 2959-2963.
Rush, M.G. and R.C. Warner (1970). Alkali denaturation of covalently closed circular duplex deoxyribonucleic acid. J. Biol. Chem. 245, 2704-2708.
Sasisekharan, V., N. Pattabiraman, and G. Gupta (1978). Some implications of an alternative structure for DNA. Proc. Natl. Acad. Sci. USA. 75(9), 4092-4096.
Sebring, E.D., T.J. Kelly Jr., M.M. Thoren and N.P Salzman (1971). Structure of replicating Simian Virus 40 deoxyribonucleic acid molecules. J. Virol. 8, 478-490.
Shure, M. and J. Vinograd (1976). The number of superhelical turns in native virion SV40 DNA and Minicol DNA determined by the band counting method. Cell 8, 215-226.
Stettler, U.H., H. Weber, T. Koller and Ch. Weissmann (1979). Preparation and characterization of form V DNA, the duplex DNA resulting from association of complementary, circular single-stranded DNA. J. Mol. Biol. 131, 21-40.
Strider, W., M.N. Camien and R.C. Warner (1981). Renaturation of Denatured, Covalently Closed Circular DNA. J. Biol. Chem. 256, 7820-7829.
Strider, W. and R.C. Warner (1971). Denatured replicative form and complex DNA of fX174: isolation and renaturation. Fed. Proc. Fed. Amer. Soc. Exp. Biol. 30(2), 1053.
Strider, W. (1971). Denatured replicative form and complex DNA of fX174: Isolation, renaturation, and sedimentation properties. Ph.D. Thesis, Department of Biochemistry, New York University School of Medicine, 550 First Avenue, New York, N.Y. 10016, U.S.A.
Travers, F., A.M. Michelson and P. Douzou (1970). Conformational changes of nucleic acids in methanol-water solutions at low temperature. Biochim. Biophys. Acta 217, 1-6.
Vinograd, J., J. Lebowitz, R. Radloff, R. Watson and P. Laipis (1965). The twisted circular form of polyoma viral DNA. Proc. Natl. Acad. Sci. USA. 53, 1104-1111.
Vinograd, J., J. Lebowitz and R. Watson (1968). Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in Polyoma DNA. J. Mol. Biol. 33, 173-197.
Wang, A.H.J., G.J. Quigley, F.J. Kolpak, J.L. Crawford, J.H. Van Boom, G. Van Der Marel and A. Rich (1979). Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680-686.
Wu R. and T.T. Wu (1996). A novel intact circular dsDNA supercoil. Bull. Math. Biol, 58(6):1171-1185.
Zimmer, C. and G. Luck (1974). Conformation and reactivity of DNA. IV. Circular dichroism studies of salt-induced conformational changes of DNAs of different base composition. Biochim. Biophys. Acta 361, 11-32.
Meaning of Z-DNA
Biology Dictionary
Definition:
A region of DNA that is "flipped" into a lefthanded
helix, characterized by alternating purines and pyrimidines, and
which may be the target of a DNA-binding protein.
http://www.hyperdictionary.com/dictionary/Z-DNA
http://www.tulane.edu/~biochem/nolan/lectures/rna/frames/zdnatxt.htm
(top view hex?!)
http://www.imb-jena.de/image_library/DNA/DNA_models/Z-DNA/zdna.html

http://www.albany.edu/~achm110/abztopview.html
NOTE B-DNA <> Z-DAN: pent <> hex as in icosa.. (also
like jitterbug effect?)
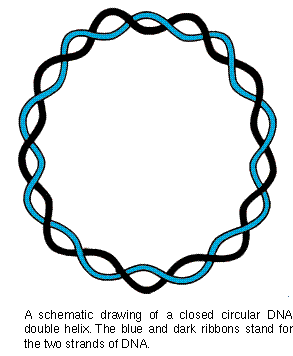
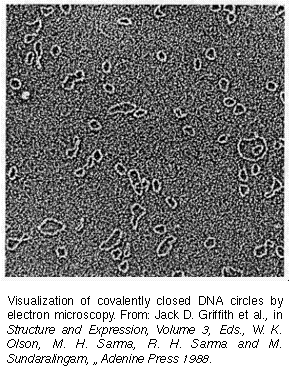
DNA Rings: Page 1
From the material so far presented there is no reason for you
to suspect that the DNA double helix to be anything other than
a long molecule, whethr bent or not, with two free ends. This
was the view until the early 1960's when electron microscopists
began examining the DNA of certain cancer viruses. To their immense
surprise, the microscopists found that some of the viral DNA's
were in the shape of closed rings (diagram). In the circular DNA
the two strands wind around each other as in a double helical
structure, but the ends are closed.
The appearence of DNA as closed circles introduces additional complexity in our visualization of DNA structure. These are not one-dimensional closed circles; but three-dimensional chains which wind ariund each other; these cricles can collapse like a ring made of a rope and wind on each other forming supercoils as illustrated below:
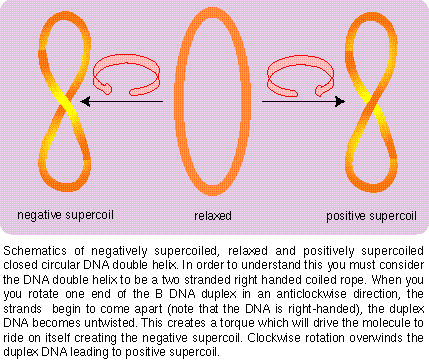
Copyright © Ramaswamy H. Sarma 1996
DNA supercoils continued continued on the next page
http://www.albany.edu/~achm110/
NOTE from
Frank- also circular dna could very well be arranged to X or Y
?!