
assembled by Dan Winter.. shared in care of the Implosion Group.. Netherlands, September 2002





|
|
Intro -prelude from Dan Winter: Academic Paper - CIRCULAR DNA - from Ken Biegeleisen.
It is a pleasure to share this RINGING DNA material - thanks to Ken Biegeleisen - who has formalized and represented his paper here on TOROIDAL or CIRCULAR DNA (relating to the concept of 'Braiding' DNA) - with academic references.
Misconception ONE: I am going to get spiritual and cosmic and immortal (biologically sustainable) when divine force (the astrology of charge?) adds '12 strands' to my DNA.
More the Truth: IF you choose the hygiene to eat, and live in, and dance in, and HAVE BLISS in-- - LIFE FORCE and therefore CHARGE - THEN your DNA may absorb that charge by enveloping itself in the recursive braid which ENSOULS - (implodes). The addition of strands which appear to happen when you braid thread into string into rope, is because of braiding (the answer lies 'folded in an envelope' - see pics below). Braiding your DNA is something you do and learn by choice - to absorb 'spin' (charge) - not something 'GOD' does for you. Only the strength of will growing in the sun-shine of shareable and embedable intent - is sustainable (and therefore able to be 'saved').
reference pictorial aritcles:
Letter to the UN on Genetic Engineering as Coherence LOSS now Measureable.
Misconception TWO: The compression spin map called "Alphabet" on the side of the "LORD OF THE RING" - RING - is too powerful for anyone to hold.
More the Truth: The 'RINGing' which causes thoughts / emotions to become objects is the implosive nature of DNA which when braided recursively by BLISS compresses charge in the only process (charge compression) which turns light (ether) into matter. (and DNA into its real destiny as a gravity making star bender) .The index to those possible angles of (Golden Ratio) compression is precisely the equation for the origin of most alphabets of Earth - specifically including Hebrew and Arabic... for the specific reason that only those biological structures which USE that symmetry discipline get to be sustainable (immortal).
Misconception THREE: DNA controls our destiny.
More the Truth: DNA is a shapeshifting mag worm which merely bends itself as a wave around the fields we form by bending the magnetism we call emotion and sacred space. It is a relatively passive mechanical linkage device designed to helpfully slow down the rate at which emotions become objects- so that we FEEL and understand the connection between CREATION and what we just now felt. Once the need for slowing down that response of stars to feeling, no longer exists - then DNA is replaced by its own etheric essence (superluminal compression field) - and we can bend light by implosion directly (only love bends the light / embedding).
If we choose consistent spin-dense and sustainable magnetic environments - then DNA responds by adding that inertia to our life force - so that eventually we gather the inertia to properly create stars and become suns ourselves. Otherwise we die a death which is permanent. DNA is merely a biofeedback device to tell us when we have consciously chosen the CHARGE and LIFE FORCE - which makes us sustainable. Short-circuiting the function of DNA by changing the spin links we call codons mechanically is not only un-sustainable and destructive - but it fundamentally ignores the design of nature - which was that DNA help us take responsibility for the SHAPE OF MAGNETISM - and so learn that bending light into shape is how we too may create stars.
Readers of the Dan Winter web site are accustomed to various speculations about the physical, mathematical and spiritual significance of the world-famous Watson-Crick "double-helix" structure of DNA, and it is therefore with some trepidation that I "rain on the parade", by showing why the double-helical structure-pretty picture that it is-is nevertheless probably not the structure of DNA in any living organism.
The primary "business" of DNA is replication. DNA carries the "codes" for the structural proteins and enzymes which give an organism its characteristics, and these codes must be passed from generation to generation.
What exactly gets passed? The codes consist of sequences of nitrogenous bases: adenine, thymine, cytosine and guanine, usually just abbreviated A, T, C, and G. These 4 "letters" are the "alphabet" of genetics, much akin to the binary code which lies at the root of all computer language. Just as a computer ultimately "spells" every program with just two numbers ("0" and "1"), likewise, the code for every structural protein and gene in the human body is "spelled" with an alphabet of only 4 bases in various combinations: A, T, C, and G.
A "gene" is therefore a linear string of bases, like a string of pearls. The bases are held together by a backbone made of sugar and phosphate. In order for the cell to execute the so-called "genetic code", it employs an extraordinary system which reads the strings of bases, and converts the information into corresponding strings of amino acids, which are the building blocks of protein. The fact that a string of bases in DNA codes for a corresponding string of amino acids in protein is sometimes called the "Central Dogma of Molecular Biology" (G-d help us! -- Is this really what we need in the post-Copernican world?).
So much for the genetics. The manner in which the bases are organized on the sugar-phosphate backbone was deduced by Watson & Crick in 1953, and, regardless of what modifications are made to their structure, this deduction will remain as one of the towering milestones in the history of science, as long as science is practiced in the world.
The manner in which the entire DNA molecule is organized, however, is another question altogether. It makes no difference to genetics whether DNA is a right-handed helix, a left-handed helix, or not-a-helix at all. Genetics simply requires that there be a string of bases, the "alphabet" if you will. Whether the sugar-phosphate backbone to which the bases are attached is twisted or not is totally irrelevant.
If this is the case, why then do scientists think that DNA is twisted?
The original structural studies of DNA were done by Maurice Wilkins, who shared the Nobel Prize with Watson & Crick (Rosalind Franklin, who actually did most of the work attributed to Wilkins, was neglected, and is now a major historical figure in the fight for women's equality). Wilkins and Franklin did what's called "x-ray crystallography". This means they took pure DNA and turned it into crystals, then took x-ray pictures of the crystals.
To make crystals, totally pure DNA is required. DNA solutions are gooey, and when a glass rod is dipped into DNA and raised up in the air, the sticky liquid is drawn into a thin thread. If the thread is drawn thin enough, it will dry out and crystallize, and a picture of the crystal can be taken with x-rays. The picture looks sort of like a snowflake. Within this very intriguing geometric pattern lies hidden information about the structure of the crystal. This information must be pulled out by mathematics. The formulae involved are difficult in the extreme, and in the old days, people who actually did the math (called "Fourier Transformation") were held somewhat in awe by other scientists. Nowadays, however, all the mathematical "leg work" is done by computers, and the process has been largely de-mystified.
The picture of DNA which emerged from the Wilkins x-ray studies was the double-helix, and very few people, including disagreeable people like myself, have ever had any reason to doubt that the structure of DNA in artificial crystals is a right-handed double-helix. But is that the structure of DNA in cells? Who can say?
Humans, insofar as our cell type is concerned, are eukaryotes, meaning that our cells are of the type referred to as "higher cells". The "lower cells" are called prokaryotes, and include, for example, bacteria.
Now, humans are not the only eukaryotes. All "higher life forms", both animal and vegetable, are eukaryotes. This means all plants, even including some unicellular plants such as mold and yeast, and all animals, even including some unicellular life forms such as paramecium or amoeba.
It is a known fact that the DNA of all eukaryotic cells is invariably bound to proteins, called histones. Here's an interesting fact for you to contemplate: The histones are exactly the same in all "higher" life forms. That means that the histones of human cells are the same as the histones of the pea plant and the amoeba. Doesn't this mean that histones are critically involved in chromosome structure? Of course it does. There's no such thing as "pure" DNA in cells! In the real world, in life, all DNA is bound to protein. There are no exceptions to this rule.
The structure of pure DNA is said to be known down to the angstrom level, and if you don't know what an angstrom is, let me tell you that it's very, very small. In other words, the structure of DNA is known down to the minutest detail.
Also, the amino acid sequences of the histones are long-since determined.
But the structure of the DNA-histone complex, called chromatin, is not known. Does this strike you as odd? It certainly strikes me as odd. After all these years, with all the importance attached to gene engineering, even after completion of the "Genome Project", the structure of human chromatin remains unknown? Incredible!
Might it be that the histones simply can't be made to fit on a "double-helix" because the "double-helix" is not the structure of DNA in cells?
As we started to say above, the primary "business" of DNA is replication. In E.coli, a common bacterium, the replication cycle can be as short as 20 minutes. This is called the "log phase" of growth, because the number of bacteria in the culture doubles every 20 minutes, giving rise to a logarithmic increase in the bacterial count.
In the log phase, the daughter cells actually begin to split into two before they have finished separating from one another! Now, that's fast!
In 1963, an Australian researcher named John Cairns took a picture of the E. coli chromosome which found its way into every textbook of that period. The picture was called an "autoradiograph", meaning that E. coli bacteria were grown on a radioactive medium, and the cells were killed and split open over a photographic plate. That plate was put in a dark room for a few months, at the end of which the tiny quantity of radiation in the chromosomes gave rise to pictures. These pictures were examined under the electron microscope.
What was the structure of the E. coli chromosome which was revealed by these autoradiographs? It was circular. Subsequent to Cairns' great discovery, it was found that nearly all the DNA which can be isolated as a single intact chromosome is also circular. This includes all bacterial DNA which has been examined, all plasmid DNA from within eukaryotic cells, and most viral DNA.
Hmmm.... that's interesting. How does a circular chromosome replicate? The two strands of a circular double-helix are LOCKED TOGETHER. If you can't imagine that clearly, I'll show you a picture:

Figure A above shows a "classic" right-handed circular double-helix (B shows a "superhelix", which we won't have time to discuss just now).
If you can't clearly envision the linkage (called "topological linkage") between the two strands in Figure A, then please think of the links of a chain. Are the links of a chain locked together? Of course they are.
How is a chain made? You can make a short 2-link chain as follows: First you take a metal rod, and close it in a circle. Then you take a second rod, pass it through the circle, wrap it around once, and close it into a second circle. Are the two links of this chain locked together? Most assuredly.
To make DNA you take one single-stranded circle of DNA, pass a straight rod of DNA through it, and wrap it around, not once but thousands of times, then seal it shut. Are the two strands locked together? You'd best believe they are. See again Figure A above.
So, if the business of DNA is replication, and if the strands must separate at replication time, and if they are indeed locked together, then how do they separate when the cell divides? Good question!
Shortly after Cairns' great discovery of circular DNA, a major DNA structural symposium was held at Cold Spring Harbor, the "Mecca" of molecular biology. The subject of DNA replication, in the light of the recent discovery of circular DNA, was indeed featured. How prominently was it featured? Not very prominently. It was briefly mentioned in one paper, are largely glossed over.
According to the scientists at the Cold Spring Harbor Symposium, how did the strands of DNA get apart during replication? The answer they gave was a "swivel". What's a "swivel"? It's a hypothetical point, somewhat comparable to an automobile universal joint, around which the strands of DNA can untwist and re-twist during the process of unwinding of all those thousands of twists, and the re-winding of the twists back in the next generation.
The swivel was thought of as an enzyme which inserted itself into the chromosome, breaking it open, and "holding on" to the DNA strands as they whirled about, trying to unwind.
What's wrong with this picture? First of all, there was no "swivel" known! It was just a guess. What about the possibility that the hypothetical "swivel" was not needed, because DNA did not have the Watson-Crick structure in living systems, but rather some alternative un-twisted structure whose strands were free to separate at will? That possibility was not even mentioned. It had evidently been determined that, even at that early date, the "double-helix" was a "sacred cow", and could not be criticized. Therefore (or so the false logic went), since DNA had to be a double-helix, and since the strands had to separate during replication, there must have been a "swivel".
What had actually happened is quite clear in retrospect. Copernicus reduced man's arrogance by removing us from the center of the universe, and placing us in a measly little planet which revolved around one of billions of stars. The "double-helix", however, put us back, squarely in the center of the universe, since we would now "create life", and become "gods". What nonsense!
But the idea was compelling to many. It was compelling enough that, for the last 40 years, almost everyone who has criticized the "double-helix" has been miserably maltreated, and rejected from the mainstream of science.
It was quickly determined, after the announcement of the mythological "swivel", that the E. coli chromosome would have to be spinning at 6,000 rpm every minute of its life if it was to unwind all those twists every 20 minutes. This is too ludicrous for words. The E. coli chromosome is something like a millimeter long when stretched out, which is an immense length of DNA to stuff into a cell so small you can't even see it without a microscope. The very idea that all this DNA is whirling like an airplane propeller at 6,000 rpm is a joke.
"No problem". The helicists simply informed us that DNA only replicates a little bit at a time, so that only one small portion of the chromosome is actually spinning at any given moment. What evidence was there that DNA replicated this way? None. It was proposed, without evidence, to save the "swivel", which itself was proposed, without evidence, to save the "double helix". This is not science.
In 1976, an Australian researcher named Gordon Rodley, fed up with the obvious shenanigans going on with DNA, published an alternative structure, which is probably the correct one. In the Rodley structure, called the "side-by-side" structure (usually just called "SBS"), the two strands of DNA lie parallel to one another, undulating a bit to the right, then a bit to the left, but never making a whole twist. Here's a very over-simplified schematic of SBS, alongside another, less likely structure; both being examples of DNA structures in which the strands are not locked together:
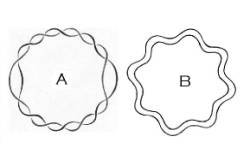
Figure A shows a totally hypothetical DNA structure which is "topologically equivalent" to the SBS structure, but in which the entire top half of the chromosome is a right-handed helix, and the entire bottom half is a left-handed helix. This model can be made instantly from two rubber bands by simply twisting them together. If you allow your rubber band model to "replicate", the two "strands" will separate as soon as you let go of them, because, topologically speaking, there were never really twisted together in the first place (every RH twist on top is canceled-out by a LH twist on the bottom). This structure, however, is highly unlikely to be found in nature.
Figure B shows the SBS structure, which is very likely to be found in nature, including in your own cells. What this drawing attempts to show is that the helix twists alternately a little to the right, then the left, etc., throughout the chromosome. As in Figure A, the net number of twists is actually 0. At replication time, the strands fall apart without having to untwist, because, topologically speaking, there really weren't any twists in the first place.
Rodley's structure (Figure B above) was published in the Proceedings of the National Academy of Science, one of the world's most rigorously peer-reviewed journals. How this got past the editors I'll never know, since Rodley remains, to this day, the only author whose work in this needlessly-controversial area has been accepted without a fight.
In 1978, Prof. Robert Chambers, the chairman of the biochemistry department at New York University School of Medicine, discovered by accident that the separated single-strands of the circular chromosome of the virus "phiX174" can spontaneously re-combine to give normal double-stranded circular DNA. This would be impossible if normal double-stranded circular DNA was a helix, because that would have required something like this:

In this drawing, a single-stranded circle is depicted as (1) being broken open, (2) wrapping itself around an intact circle, and (3) sealing itself shut again. The breaking open in step (1) is not a problem, because DNA can certainly break. The wrapping around the other strand in step (2) is not a problem because DNA is known to do that. But the sealing of the broken strand shut in step (3) is a BIG problem, because that requires a sealing enzyme, and no such enzyme was present in Prof. Chambers' single-stranded circular DNA preparation.
Is there another way to explain this data? Of course, but you have to be willing to believe that DNA is not a helix. It goes like this:

Here, the two circles are being shown simply coming together to form un-twisted double-stranded DNA. (Actually, it would be slightly twisted, but only as per the SBS structure two figures above this one).
Why has this simple concept been resisted so strongly? Because is was also discovered, not long after the discovery of circular DNA itself, that the two strands of circular DNA, when "melted", do not separate, which they ought to do if they are not locked together.
"Melting" doesn't mean literal melting, as when you heat butter in a frying pan. It means subjecting the DNA to conditions which promote strand separation. The first "melting" experiments were on fragmented linear DNA, and the procedure employed was simple boiling. At the temperature of boiling water, the double-stranded structure broke down, releasing single-stranded linear DNA. If the solutions were cooled slowly, the double-stranded form would, however, re-appear.
On the other hand, when circular DNA is boiled the strands do not separate. Moreover, at pH 13 (very, alkaline, like lye), where normal double-stranded DNA structure is impossible to maintain, the strands of circular DNA still do not separate! What's wrong?
I began working on this problem in the early 1970's. It took me years to solve the whole puzzle. In 1974 I submitted my first theoretical article on the subject to the Journal of Molecular Biology, which rudely rejected it. It took 28 years to get this article into a peer-reviewed scientific journal, but it was finally published a few months ago, in the Bulletin of Mathematical Biology (Biegeleisen, K. Topologically Non-linked Circular Duplex DNA, Bull Math Biol 64:589-609, May 2002).
The content of the article is a bit too long and drawn-out for the present discussion. Suffice it to say that the explanation for the well-documented fact that the strands of circular DNA do not separate under "melting" conditions is that the circularization of the DNA imparts novel and unexpected topological properties to the chromosome; properties which cause it to behave markedly differently from linear DNA under the same circumstances.
In particular, at high pH, where the single-strands of linear DNA fall apart, the single strands of circular DNA remain associated in a new, multi-strand, alkali-stable complex. The structure for this complex is similar to the structure originally proposed for all DNA by Linus Pauling, just months before the famous paper by Watson & Crick hit the press. Pauling, in his day, was considered to be the world's greatest living authority on the chemical bond, and the odds-makers had picked him to win the "race" to determine the structure of DNA. He considered a triple or quadruple helix with the phosphate groups pointing inward to be the most logical structure for DNA, based on the chemistry only (i.e., without reference to genetically-specific base-pairing, which had not yet been discovered).
When Watson & Crick published the "double helix", it looked as if Pauling had "lost the race". Now, however, almost exactly 50 years later, we can see that maybe he didn't lost it entirely.
If circular duplex DNA really has the SBS structure, or one similar to it, then it ought to be possible to separate the component single-stranded circles, without breaking either one open. This was not accomplished for many years.
Proof that the strands of circular DNA can indeed be separated was finally published in 1996, by a brilliant researcher named Tai Te Wu, with the help of his son (Wu R. and T.T. Wu, 1996. A novel intact circular dsDNA supercoil. Bull Math Biol, 58(6):1171-1185).
Wu's innovation was to isolate double-stranded chromosomal DNA from a plasmid under conditions where a great deal of a substance called "RNA" was bound to only one of the two strands of the DNA chromosome. The bound RNA changed the physical characteristics of the strand it was bound to, so that Wu was able to separate the two chromosomal strands by prolonged gel electrophoresis (a procedure in which DNA's of different physical characteristics can be separated into distinct bands in a jello-like substance to which an electric current has been applied). Wu repeated the work with a second plasmid which had the two strands marked by the intentional insertion of marker sequences by genetic engineering, and proved, unequivocally, that the two bands in the gel were indeed the two single-strands of the original plasmid chromosome, respectively.
The conclusion is that DNA is not a helix, and it's really been apparent all along to many people. The failure of the two strands of circular DNA to separate under "melting" conditions made it possible for the scientific community to evade this obvious conclusion for many years, but the days of the "double-helix" as the presumed structure of DNA in living system are, in all probability, numbered.







